Evaluating the innovation capability of DP2
Evaluating innovation capability of the algorithm using different version of a curated database for comparison
In the “data revolution era” that we are facing it’s not so difficult to find ourselves overwhelmed by data, with the risk that a possible resource became an obstacle. For this reason, the rise of AI has been so crucial in the last few years and probably it will be more and more in the future. It was this context that brings to the foundation of TheProphetAI start-up bearing in mind a precise question: how can we better use the knowledge already present in the field of life science to provide a boost to the research?
The answer materialized in GeneRecommender, a free platform powered by a DNN trained considering the co-citation of genes in the Pubmed scientific literature and so capable of building a representation (embedding) of all human genome with a broad perspective scientifically based. The algorithm accepts a set of genes and produces as output a new one, ranked by a similarity score, and with possible correlations with the input.
It’s not the scope of this article to explain in detail the architecture of the net or the evaluation step performed (for that it’s available a scientific paper available in preprint at https://arxiv.org/abs/2208.01918) but test the capability of the system to recommend knowledge not already established in the scientific community.
To accomplish this task, an open-access and well-curated knowledge base relative to biological pathways, Reactome, was selected. The idea was to compare two different versions of this database to isolate discoveries in the field and check if the AI algorithm, trained with an older knowledge base, can predict some of these breakthroughs.
The first test was carried out using a training dataset composed of articles published before march 2021 and giving as input to the DNN the gene sets constituting the pathway in the Reactome version of the same period (version 76, 3/23/2021). The output of the net was cut to the first 50 genes ranked by similarity with input and the number of suggestions that turned out to be right was checked. As ground truth was considered the differences between the aforementioned version of Reactome and one subsequent year (version 80, 3/23/2022).
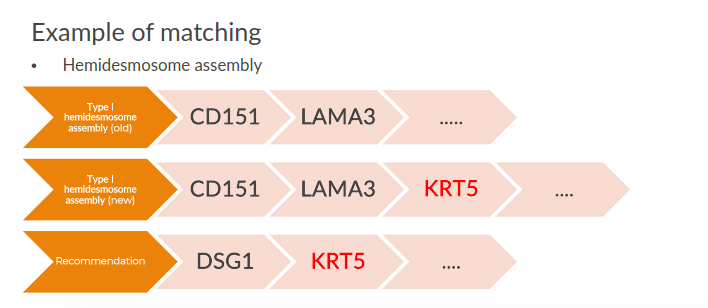
In that interval of time, 657 pathways were been updated in Reactome with the addition of some genes and 24% of them have at least a matching in the AI predictions.
Considering the genes, 902 distinct genes have been introduced in the pathways and 9% of them have also a correct matching in the predictions.
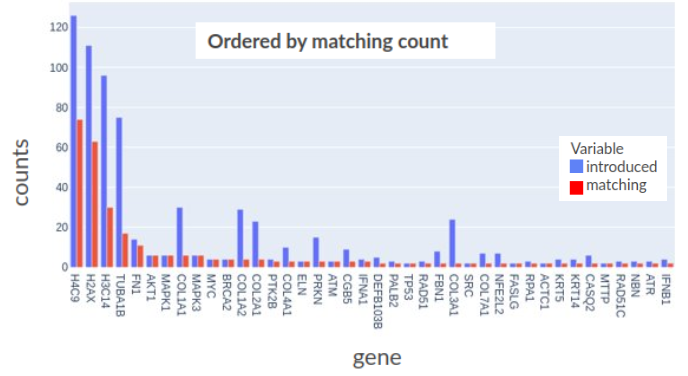
In the below figure are plotted the counts of introductions or matchings per gene, could be observed that for some of them, the number coincides, meaning that every time they are added to a pathway a recommendation predicted it.
The same analysis was repeated using different versions of the Reactome database (December 2019, version 69 versus March 2021, version 76) And this time the training dataset had been cut off to June 2017. In this way, with two years of lag between the training source and the data used as input to get predictions, the risk of having knowledge already established but not yet updated in Reactome is minimized. The results of this test can so be considered a measure of the capability for the system to produce innovative suggestions.
In this case, the pathways updated are 430 and 47% of them have at least a matching. The distinct genes added are instead 1846 and 9% of them have matching in the predictions. It’s interesting to notice how the percentage of pathways with a match nearly doubled considering a broader interval (2019 vs 2021 respect 2021 vs 2022), as if increasing the time given to the research process can produce more matchings with the AI predictions. At support to this hypothesis are the data obtained considering an even larger interval between Reactome versions, 2017 vs 2021: 54% of pathways with match and 12% of distinct genes with a match. It’s not to be excluded that the AI predictions contain even more correct suggestions that will be added in the future.
Anyway, this sort of analysis proved that the predictions produced by the AI contain some innovative suggestions that could be helpful for the research. Maybe not all the suggestions are correct but the aim of this Neural Network was not to substitute the researchers but instead be a tool that he could use to gain a different perspective and ask himself the right questions.
It’s not an easy task to evaluate a recommender system powered by AI, even more, if it aims to produce an innovative recommendation for researchers but this had proven to be an interesting approach.
We think that we are on the verge of a revolution in medical research where AI will play a pivotal role, Generecommender is a step in that direction. Like any innovative technology, not all the steps define a new road, but surely any of them map the field to find the way.
